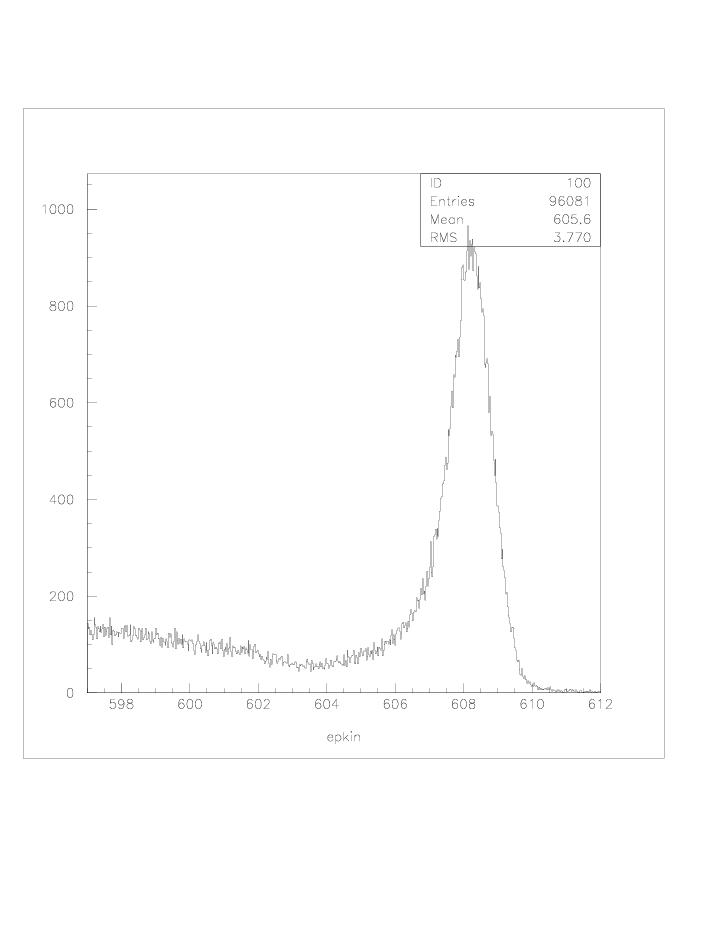

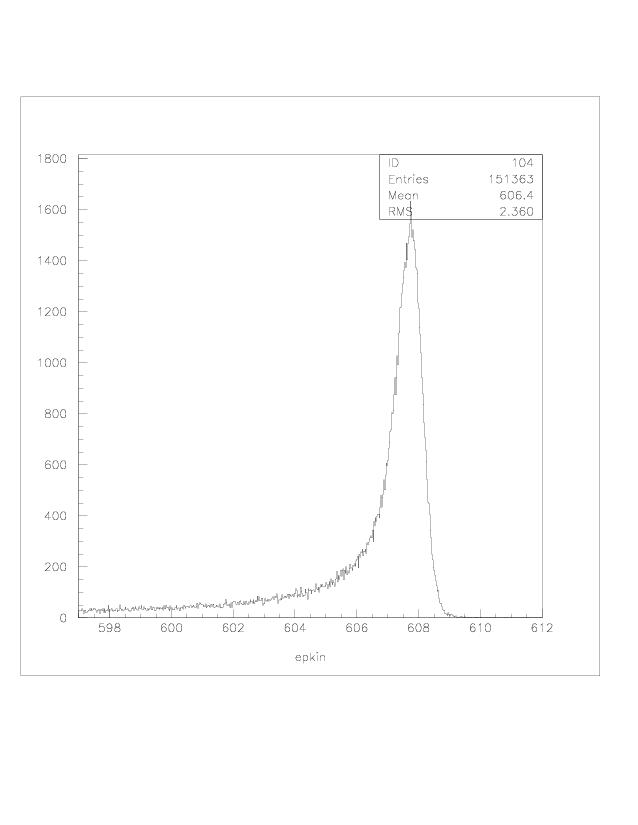
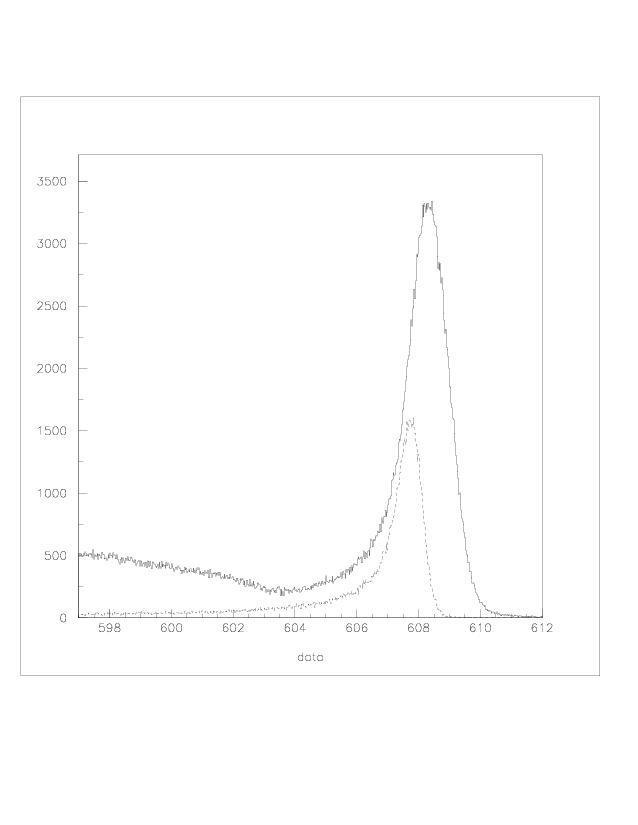
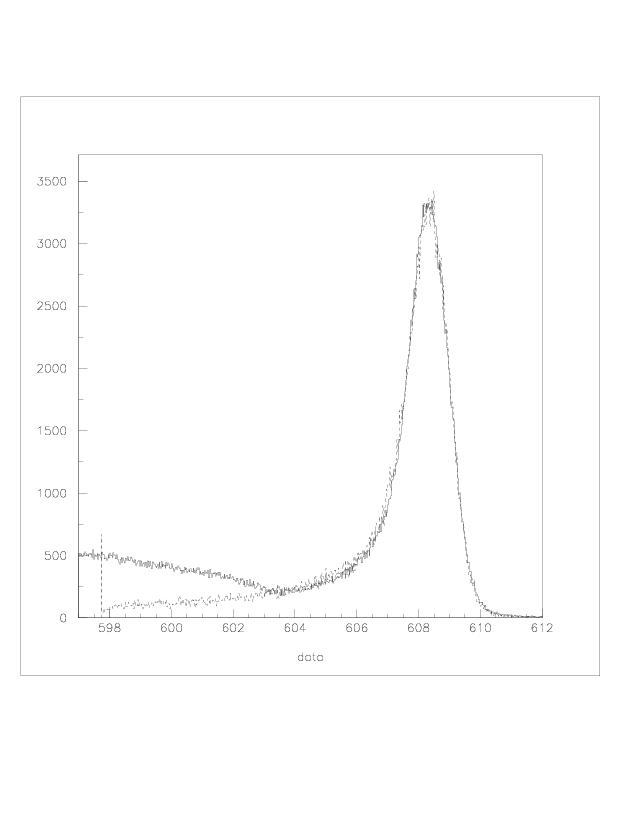
| q2 |
644 |
842 |
1257 |
1956 |
2903 |
4032 |
| 1.29 |
DH, 20.17 (625,640) |
|||||
| 1.31 |
ER, 15.49 (821,836) |
|||||
| 1.59 |
DH, 22.46 (621,636) |
|||||
| 1.88 |
DH, 24.46 (618,633) |
|||||
| 1.90 |
ER, 18.72 (817,832) |
|||||
| 2.26 |
DH, 26.96 (615,630) |
|||||
| 2.79 |
DH, 15.19 (1222,1237) |
|||||
| 2.91 |
DH, 30.77 (611,626) |
|||||
| 3.7 |
DH, 34.97 605,620) |
|||||
| 4.56 |
DH, 39.17 (599,614) |
|||||
| 4.99 |
DH, 41.20 (597,612) |
ER, 13.05 (1909,1924) |
||||
| 5.01 |
ER, 31.03 (797,812) |
DH, 20.57 (1207,1222) |
||||
| 10.1 |
ER, 45.67 (760,775) |
|||||
| 11.01 |
DH, 31.16 (1168,1183) |
|||||
| 14.94 |
ER, 15.37 (2788,2803) |
|||||
| 14.98 |
ER, 36.96 (1141,1156) |
|||||
| 15 |
ER, 57.91 (726,741) |
|||||
| 19.06 |
ER, 42.46 (1113,1128) |
|||||
| 19.37 |
ER, 12.56 (3880,3940) over 300 channels |
|||||
| 19.39 |
ER, 68.42 (696,711) |